
 |
|
|
Главная --> Промиздат --> Map principle - like GRASS modules in general - assumes that the user needs the current resolution, not the initial resolution of the map layer. The current interface is supporting raster and site data. Work on an interface for vector data is ongoing. Besides the base package of R it is useful to install also the following contributed extensions: akima, fields, geoR, grid, lattice, MASS, scatterplot3d, spatial, and stepfun (available from the R Web site). Additional packages in terms of spatial-temporal analysis are focused on autocorrelation, spatial point patterns, time series or wavelets. Note that in this section we omit the \ character for long lines as it is not allowed in R. Long, broken lines are indicated by the indent in the next line. Installation of the R/GRASS interface. The installation of the R/GRASS interface9 is very easy and can be done by a single command (you probably have to be user root for this). If your computer is connected to the Internet, you can install packages within an R session. Start R and launch the command: install.packages( GRASS ) This will download the latest version of the selected package, unpack, compile and install it. From time to time the installed extensions should be checked for updates. The following command will download the R package list, compare to the local installation and upgrade installed packages if new versions are available: update.packages() Offline, you can install extra packages on command line: R CMD INSTALL GRASS 0.2-13.tar.gz As the interface is maturing, the version number is subject to change. Also the path to R depends on your local installation. To start a quick exploration of standard R, simply run: demo () demo(graphics) The function q() finishes a R session. When leaving R with q() you will be asked: Save workspace image? [y/n/c]: . If answering y, the objects are stored within the local directory into the hidden file .RData. When launching R next time in this directory, the objects will be read into the system and you can continue with your work. As mentioned above, contributed packages extend immensely the functionality of GRASS in conjunction with the R/GRASS interface. To find out which packages are already available on your system, type within R: You can load an installed package by entering its name as a parameter for this function. Some packages also provide examples for their functions. We try an example for fitting a trend surface, the function is provided by library spatial: library(spatial) example(surf.Is) ?surf.Is The ? command displays the functions help text (leave with q). Help pages are also stored in HTML format, you can open a HTML browser with: help . start () The pages provide package explanations and a local search engine. When using R in batch mode you can develop GRASS scripts, which are directly utilizing R functionality in GRASS user environment. In Section 13.2.3 we show sample scripts. 13.2.1 Spearfish data set analysis To illustrate how to apply R to your data, we present several examples based on the Spearfish data set. As an exercise, we can generate a trend surface of the distributed pH value within the Spearfish region. After starting GRASS with Spearfish LOCATION and resetting the region to default settings, run R within GRASS: grass53 /usr/local/share/grassdata/spearfish/user1 g.region -dp Within R the R/GRASS interface is loaded as follows: library(GRASS) G <- gmetaO #show loaded environment: class(G) summary(G) By this, we have loaded the library of interface functions and initialized an object named G that includes metadata about the location. This object, like many in R, has a class grassmeta- which is used for selecting functions that are applied to the object, for example, summary() or plot(). The class of the object, which is the first argument to a generic function, passes the object on to a function specifically for that object class, so function summary(G) gives a summary appropriate for objects of class grassmeta. A little later, we will see summary() used on numeric data, and the plot() function used with G as the first argument. Now we are ready to perform geospatial analysis of GRASS raster and site data. As a prerequisite to generate the trend map of pH values with R, the pH values map is read into R environment (with c(F) category import set to False): 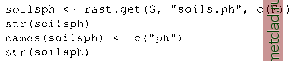 The function rast.getO reads the map soils.ph from GRASS and stores it into the R object soilsph. The str() function shows a brief view of the structure of the soilsph object. It starts with NA (no-data) values because the GRASS soils.ph map contains NULL values at the borders. For later convenience the imported variable is renamed from soilsph$soils.ph to soilsph$ph with the names() function. Univariate statistics (minimum, maximum, 1st and 3rd quartile, median, mean and the number of NAs) of the pH values can be displayed with summary() function. Up to now the recently imported map stored in data object soilsph contains only the data values, but not the related coordinates. To complete it, we generate a so-called data frame which allows us to store associated variables in one object. In our case, the data frame will carry east and north coordinates and the z values, which represent the ph values: summary(soilsphSph) soils.ph.frame <- data.frame(east(G), str(soils.ph.frame) system( r.report soils.ph ) north (G), soilsph$ph) Since the original soils.ph map contains cells with no soil data label (checkwith r.report) we have to set these values to NA. For further processing we rename the variables to standard naming convention. Most functions expect the variable names to be x , y and z . Note that when calling an object in R by name, the object contents will be displayed (e.g. all cell values): soilsph$ph[soilsph$ph == 0] <- NA names(soils.ph.frame) <- c( x , y , z ) summary(soils.ph.frame) soils.ph.frame$z
|